Note
Go to the end to download the full example code.
Variable Importance on diabetes dataset#
Variable Importance estimates the influence of a given input variable to the prediction made by a model. To assess variable importance in a prediction problem, Breiman[1] introduced the permutation approach where the values are shuffled for one variable/column at a time. This permutation breaks the relationship between the variable of interest and the outcome. Following, the loss score is checked before and after this substitution for any significant drop in the performance which reflects the significance of this variable to predict the outcome. This ease-to-use solution is demonstrated, in the work by Strobl et al.[2], to be affected by the degree of correlation between the variables, thus biased towards truly non-significant variables highly correlated with the significant ones and creating fake significant variables. They introduced a solution for the Random Forest estimator based on conditional sampling by performing sub-groups permutation when bisecting the space using the conditioning variables of the building process. However, this solution is exclusive to the Random Forest and is costly with high-dimensional settings. Chamma et al.[3] introduced a new model-agnostic solution to bypass the limitations of the permutation approach under the use of the conditional schemes. The variable of interest does contain two types of information: 1) the relationship with the remaining variables and 2) the relationship with the outcome. The standard permutation, while breaking the relationship with the outcome, is also destroying the dependency with the remaining variables. Therefore, instead of directly permuting the variable of interest, the variable of interest is predicted by the remaining variables and the residuals of this prediction are permuted before reconstructing the new version of the variable. This solution preserves the dependency with the remaining variables.
In this example, we compare both the standard permutation and its conditional variant approaches for variable importance on the diabetes dataset for the single-level case. The aim is to see if integrating the new statistically-controlled solution has an impact on the results.
References#
Load the diabetes dataset#
from sklearn.datasets import load_diabetes
diabetes = load_diabetes()
X, y = diabetes.data, diabetes.target
# Encode sex as binary
X[:, 1] = (X[:, 1] > 0.0).astype(int)
Fit a baseline model on the diabetes dataset#
We use a Ridge regression model with a 5-fold cross-validation to fit the diabetes dataset.
import numpy as np
from sklearn.base import clone
from sklearn.linear_model import LogisticRegressionCV, RidgeCV
from sklearn.metrics import r2_score, root_mean_squared_error
from sklearn.model_selection import KFold
n_folds = 5
regressor = RidgeCV(
alphas=np.logspace(-3, 3, 10),
cv=KFold(shuffle=True, random_state=20),
)
regressor_list = [clone(regressor) for _ in range(n_folds)]
kf = KFold(n_splits=n_folds, shuffle=True, random_state=21)
for i, (train_index, test_index) in enumerate(kf.split(X)):
regressor_list[i].fit(X[train_index], y[train_index])
score = r2_score(
y_true=y[test_index], y_pred=regressor_list[i].predict(X[test_index])
)
mse = root_mean_squared_error(
y_true=y[test_index], y_pred=regressor_list[i].predict(X[test_index])
)
print(f"Fold {i}: {score=}")
print(f"Fold {i}: {mse=}")
Fold 0: score=0.42498602106170535
Fold 0: mse=56.42055334073377
Fold 1: score=0.4576508881883167
Fold 1: mse=58.423308135994034
Fold 2: score=0.5351768182569143
Fold 2: mse=54.730052311563774
Fold 3: score=0.48890245901647933
Fold 3: mse=53.411951707073335
Fold 4: score=0.5377531849343379
Fold 4: mse=51.2067043978449
Measure the importance of variables using the CFI method#
from hidimstat import CFI
cfi_importance_list = []
kf = KFold(n_splits=n_folds, shuffle=True, random_state=21)
for i, (train_index, test_index) in enumerate(kf.split(X)):
print(f"Fold {i}")
X_train, X_test = X[train_index], X[test_index]
y_train, y_test = y[train_index], y[test_index]
cfi = CFI(
estimator=regressor_list[i],
imputation_model_continuous=RidgeCV(alphas=np.logspace(-3, 3, 10), cv=KFold()),
imputation_model_categorical=LogisticRegressionCV(
Cs=np.logspace(-2, 2, 10),
cv=KFold(),
),
# covariate_estimator=HistGradientBoostingRegressor(random_state=0,),
n_permutations=50,
random_state=24,
n_jobs=4,
)
cfi.fit(X_train, y_train)
importance = cfi.importance(X_test, y_test)
cfi_importance_list.append(importance)
Fold 0
Fold 1
Fold 2
Fold 3
Fold 4
Measure the importance of variables using the LOCO method#
from hidimstat import LOCO
loco_importance_list = []
kf = KFold(n_splits=n_folds, shuffle=True, random_state=21)
for i, (train_index, test_index) in enumerate(kf.split(X)):
print(f"Fold {i}")
X_train, X_test = X[train_index], X[test_index]
y_train, y_test = y[train_index], y[test_index]
loco = LOCO(
estimator=regressor_list[i],
n_jobs=4,
)
loco.fit(X_train, y_train)
importance = loco.importance(X_test, y_test)
loco_importance_list.append(importance)
Fold 0
Fold 1
Fold 2
Fold 3
Fold 4
Measure the importance of variables using the permutation method#
from hidimstat import PFI
pfi_importance_list = []
kf = KFold(n_splits=n_folds, shuffle=True, random_state=21)
for i, (train_index, test_index) in enumerate(kf.split(X)):
print(f"Fold {i}")
X_train, X_test = X[train_index], X[test_index]
y_train, y_test = y[train_index], y[test_index]
pfi = PFI(
estimator=regressor_list[i],
n_permutations=50,
random_state=25,
n_jobs=4,
)
pfi.fit(X_train, y_train)
importance = pfi.importance(X_test, y_test)
pfi_importance_list.append(importance)
Fold 0
Fold 1
Fold 2
Fold 3
Fold 4
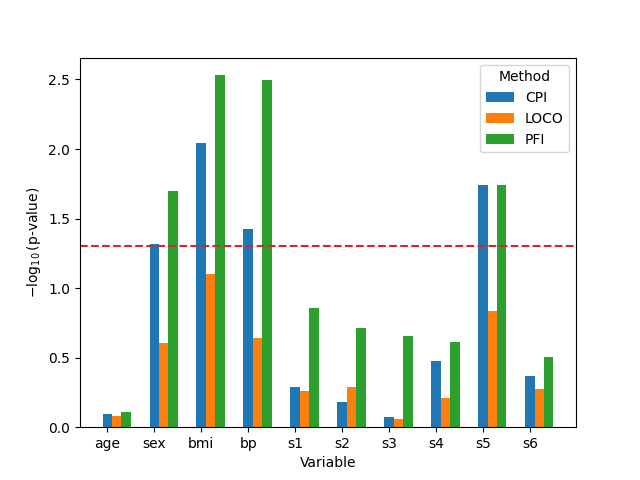
Analyze the results#
import matplotlib.pyplot as plt
import pandas as pd
from scipy.stats import ttest_1samp
cfi_vim_arr = np.array([x["importance"] for x in cfi_importance_list]) / 2
cfi_pval = ttest_1samp(cfi_vim_arr, 0, alternative="greater").pvalue
vim = [
pd.DataFrame(
{
"var": np.arange(cfi_vim_arr.shape[1]),
"importance": x["importance"],
"fold": i,
"pval": cfi_pval,
"method": "CFI",
}
)
for x in cfi_importance_list
]
loco_vim_arr = np.array([x["importance"] for x in loco_importance_list])
loco_pval = ttest_1samp(loco_vim_arr, 0, alternative="greater").pvalue
vim += [
pd.DataFrame(
{
"var": np.arange(loco_vim_arr.shape[1]),
"importance": x["importance"],
"fold": i,
"pval": loco_pval,
"method": "LOCO",
}
)
for x in loco_importance_list
]
pfi_vim_arr = np.array([x["importance"] for x in pfi_importance_list])
pfi_pval = ttest_1samp(pfi_vim_arr, 0, alternative="greater").pvalue
vim += [
pd.DataFrame(
{
"var": np.arange(pfi_vim_arr.shape[1]),
"importance": x["importance"],
"fold": i,
"pval": pfi_pval,
"method": "PFI",
}
)
for x in pfi_importance_list
]
fig, ax = plt.subplots()
df_plot = pd.concat(vim)
df_plot["pval"] = -np.log10(df_plot["pval"])
methods = df_plot["method"].unique()
colors = plt.get_cmap("tab10", 10)
for i, method in enumerate(methods):
subset = df_plot[df_plot["method"] == method]
ax.bar(
subset["var"] + i * 0.2,
subset["pval"],
width=0.2,
label=method,
color=colors(i),
)
ax.legend(title="Method")
ax.set_ylabel(r"$-\log_{10}(\text{p-value})$")
ax.axhline(-np.log10(0.05), color="tab:red", ls="--")
ax.set_xlabel("Variable")
ax.set_xticks(range(len(diabetes.feature_names)))
ax.set_xticklabels(diabetes.feature_names)
plt.show()
Total running time of the script: (0 minutes 33.766 seconds)
