Note
Go to the end to download the full example code.
Variable Selection Under Model Misspecification#
In this example, we illustrate the limitations of variable selection methods based on linear models using the circles dataset. We first use the distilled conditional randomization test (d0CRT), which is based on linear models Liu et al.[1] and then demonstrate how model-agnostic methods, such as Leave-One-Covariate-Out (LOCO), can identify important variables even when classes are not linearly separable.
To evaluate the importance of a variable, LOCO re-fits a sub-model using a subset of the data where the variable of interest is removed. The importance of the variable is quantified as the difference in loss between the full model and the sub-model. As shown in Williamson et al.[2] , this loss difference can be interpreted as an unnormalized generalized ANOVA (difference of R²). Denoting \(\mu\) the predictive model used, \(\mu_{-j}\) the sub-model where the j-th variable is removed, and \(X^{-j}\) the data with the j-th variable removed, the loss difference can be expressed as:
where \(\psi_{j}\) is the LOCO importance of the j-th variable.
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from scipy.stats import ttest_1samp
from sklearn.base import clone
from sklearn.datasets import make_circles
from sklearn.linear_model import LogisticRegressionCV
from sklearn.metrics import hinge_loss, log_loss
from sklearn.model_selection import KFold
from sklearn.svm import SVC
from hidimstat import LOCO, dcrt_pvalue, dcrt_zero
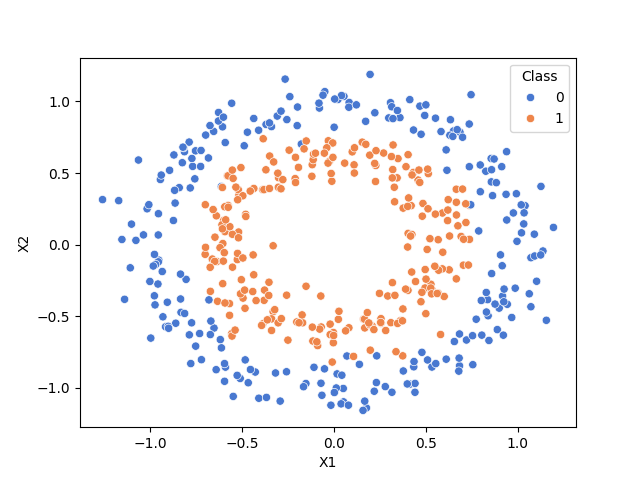
Generate data where classes are not linearly separable#
rng = np.random.RandomState(0)
X, y = make_circles(n_samples=500, noise=0.1, factor=0.6, random_state=rng)
fig, ax = plt.subplots()
sns.scatterplot(
x=X[:, 0],
y=X[:, 1],
hue=y,
ax=ax,
palette="muted",
)
ax.legend(title="Class")
ax.set_xlabel("X1")
ax.set_ylabel("X2")
plt.show()
Compute p-values using d0CRT#
We first compute the p-values using d0CRT which performs a conditional independence test (\(H_0: X_j \perp\!\!\!\perp y | X_{-j}\)) for each variable. However, this test is based on a linear model (LogisticRegression) and fails to reject the null in the presence of non-linear relationships.
selection_features, X_residual, sigma2, y_res = dcrt_zero(
X, y, problem_type="classification", screening=False
)
_, pval_dcrt, _ = dcrt_pvalue(
selection_features=selection_features,
X_res=X_residual,
y_res=y_res,
sigma2=sigma2,
fdr=0.05,
)
/home/circleci/project/.venv/lib/python3.13/site-packages/sklearn/linear_model/_coordinate_descent.py:1613: FutureWarning: 'n_alphas' was deprecated in 1.7 and will be removed in 1.9. 'alphas' now accepts an integer value which removes the need to pass 'n_alphas'. The default value of 'alphas' will change from None to 100 in 1.9. Pass an explicit value to 'alphas' and leave 'n_alphas' to its default value to silence this warning.
warnings.warn(
/home/circleci/project/.venv/lib/python3.13/site-packages/sklearn/linear_model/_coordinate_descent.py:1632: FutureWarning: 'alphas=None' is deprecated and will be removed in 1.9, at which point the default value will be set to 100. Set 'alphas=100' to silence this warning.
warnings.warn(
/home/circleci/project/.venv/lib/python3.13/site-packages/sklearn/linear_model/_coordinate_descent.py:1613: FutureWarning: 'n_alphas' was deprecated in 1.7 and will be removed in 1.9. 'alphas' now accepts an integer value which removes the need to pass 'n_alphas'. The default value of 'alphas' will change from None to 100 in 1.9. Pass an explicit value to 'alphas' and leave 'n_alphas' to its default value to silence this warning.
warnings.warn(
/home/circleci/project/.venv/lib/python3.13/site-packages/sklearn/linear_model/_coordinate_descent.py:1632: FutureWarning: 'alphas=None' is deprecated and will be removed in 1.9, at which point the default value will be set to 100. Set 'alphas=100' to silence this warning.
warnings.warn(
/home/circleci/project/.venv/lib/python3.13/site-packages/sklearn/linear_model/_coordinate_descent.py:1613: FutureWarning: 'n_alphas' was deprecated in 1.7 and will be removed in 1.9. 'alphas' now accepts an integer value which removes the need to pass 'n_alphas'. The default value of 'alphas' will change from None to 100 in 1.9. Pass an explicit value to 'alphas' and leave 'n_alphas' to its default value to silence this warning.
warnings.warn(
/home/circleci/project/.venv/lib/python3.13/site-packages/sklearn/linear_model/_coordinate_descent.py:1632: FutureWarning: 'alphas=None' is deprecated and will be removed in 1.9, at which point the default value will be set to 100. Set 'alphas=100' to silence this warning.
warnings.warn(
Compute p-values using LOCO#
We then compute the p-values using LOCO with a linear, and then a non-linear model. When using a misspecified model, such as a linear model for this dataset, LOCO fails to reject the null similarly to d0CRT. However, when using a non-linear model (SVC), LOCO is able to identify the important variables.
cv = KFold(n_splits=5, shuffle=True, random_state=0)
non_linear_model = SVC(kernel="rbf", random_state=0)
linear_model = LogisticRegressionCV(Cs=np.logspace(-3, 3, 5))
importances_linear = []
importances_non_linear = []
for train, test in cv.split(X):
non_linear_model_ = clone(non_linear_model)
linear_model_ = clone(linear_model)
non_linear_model_.fit(X[train], y[train])
linear_model_.fit(X[train], y[train])
vim_linear = LOCO(
estimator=linear_model_, loss=log_loss, method="predict_proba", n_jobs=2
)
vim_non_linear = LOCO(
estimator=non_linear_model_,
loss=hinge_loss,
method="decision_function",
n_jobs=2,
)
vim_linear.fit(X[train], y[train])
vim_non_linear.fit(X[train], y[train])
importances_linear.append(vim_linear.importance(X[test], y[test])["importance"])
importances_non_linear.append(
vim_non_linear.importance(X[test], y[test])["importance"]
)
To select variables using LOCO, we compute the p-values using a t-test over the importance scores.
_, pval_linear = ttest_1samp(importances_linear, 0, axis=0, alternative="greater")
_, pval_non_linear = ttest_1samp(
importances_non_linear, 0, axis=0, alternative="greater"
)
df_pval = pd.DataFrame(
{
"pval": np.hstack([pval_dcrt, pval_linear, pval_non_linear]),
"method": ["d0CRT"] * 2 + ["LOCO-linear"] * 2 + ["LOCO-non-linear"] * 2,
"Feature": ["X1", "X2"] * 3,
}
)
df_pval["minus_log10_pval"] = -np.log10(df_pval["pval"])
Plot the \(-log_{10}(pval)\) for each method and variable#
fig, ax = plt.subplots()
sns.barplot(
data=df_pval,
y="Feature",
x="minus_log10_pval",
hue="method",
palette="muted",
ax=ax,
)
ax.set_xlabel("-$\\log_{10}(pval)$")
ax.axvline(
-np.log10(0.05), color="k", lw=3, linestyle="--", label="-$\\log_{10}(0.05)$"
)
ax.legend()
plt.show()
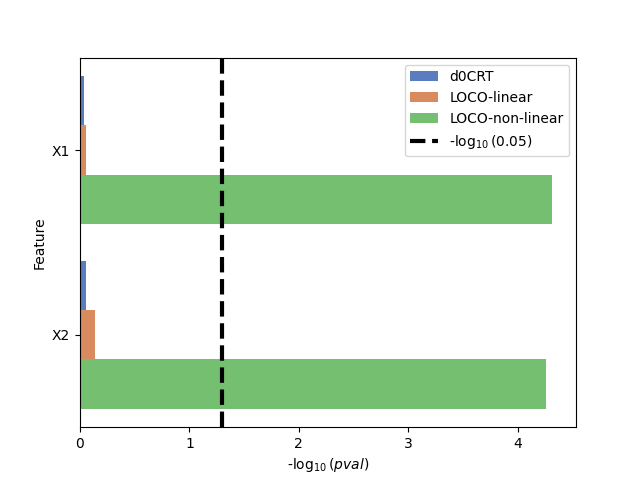
As expected, when using linear models (d0CRT and LOCO-linear) that are misspecified, the varibles are not selected. This highlights the benefit of using model-agnostic methods such as LOCO, which allows for the use of models that are expressive enough to explain the data.
References#
Total running time of the script: (0 minutes 6.047 seconds)
